Quick start with example data
Quick start with example data
To take your first steps with HiCognition, you can download example data from our dropbox location. The files located there are centered on analysis connected to sister chromatid sensitive Hi-C.
Example 1: Conformation of sister chromatids around TADs
Needed data:
G2_tads_w_size.bed| The location of TADs in G2 synchronized HeLa cellsG2.fc_1_2_3_4.wOldG2.cis.1000.mcool| Cis-sister contacts in G2 synchronized HeLa cellsG2.fc_1_2_3_4.wOldG2.trans.1000.mcool| Trans-sister contacts in G2 synchronized HeLa cells
To upload the files, follow our upload instructions for genomic regions and genomic features. The metadata settings are the following:
TADs:
Perturbation| noneCell cycle stage| G2Genome assembly| hg19Sizetype| IntervalValueType| DerivedMethod| HiC
Cooler files:
Perturbation| noneCell cycle stage| G2Genome assembly| hg19ValueType| InteractionMethod| HiCNormalization| ICCF
The .mcool files are quite big, and you should expect them to take a few minutes to upload (depending on your internet connection).
Once the files are uploaded, start preprocessing the two 2D features at TAD-boundaries by following our preprocessing guide. Once preprocessing is finished, you can create a widget collection by clicking on the  at the bottom right of the HiCognition start page. This will create an empty widget collection:
at the bottom right of the HiCognition start page. This will create an empty widget collection:

By clicking on the region-controls, you can select your TADs as a region. Once that is done, hover over the center +-icon to reveal the widget-type menu:
Once you select the 2D-average widget, you can select one of the two 2D features you added. For example, if you select the cis-sister contacts, your widget collection should look like this:
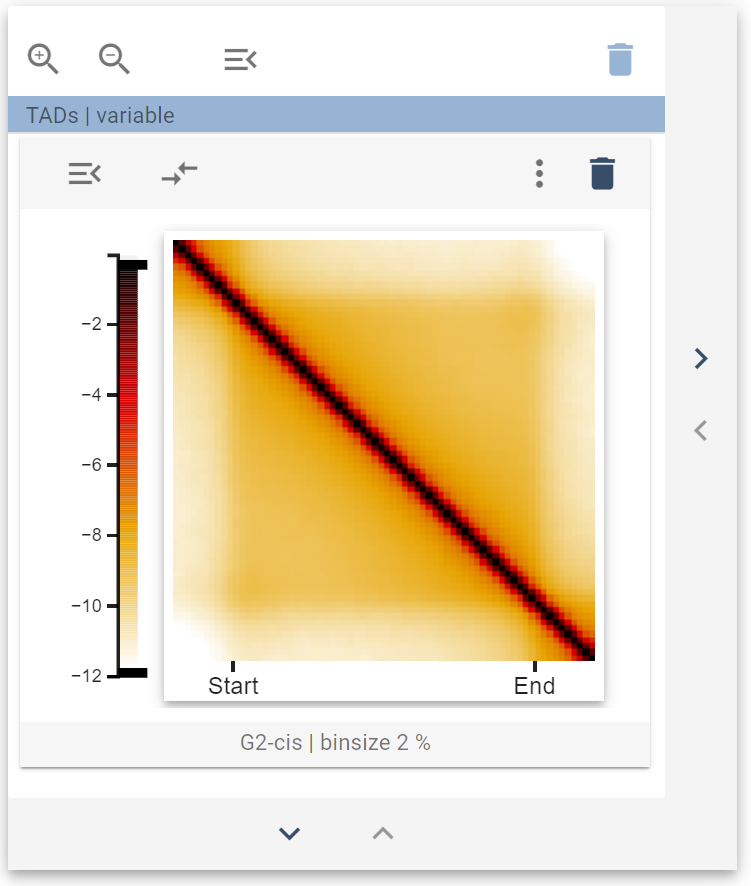
This picture represents the average behavior of cis-sister contacts around TADs. You can then resize your widget collection to add trans-sister contacts as a second widget. The resulting collection should look like this:
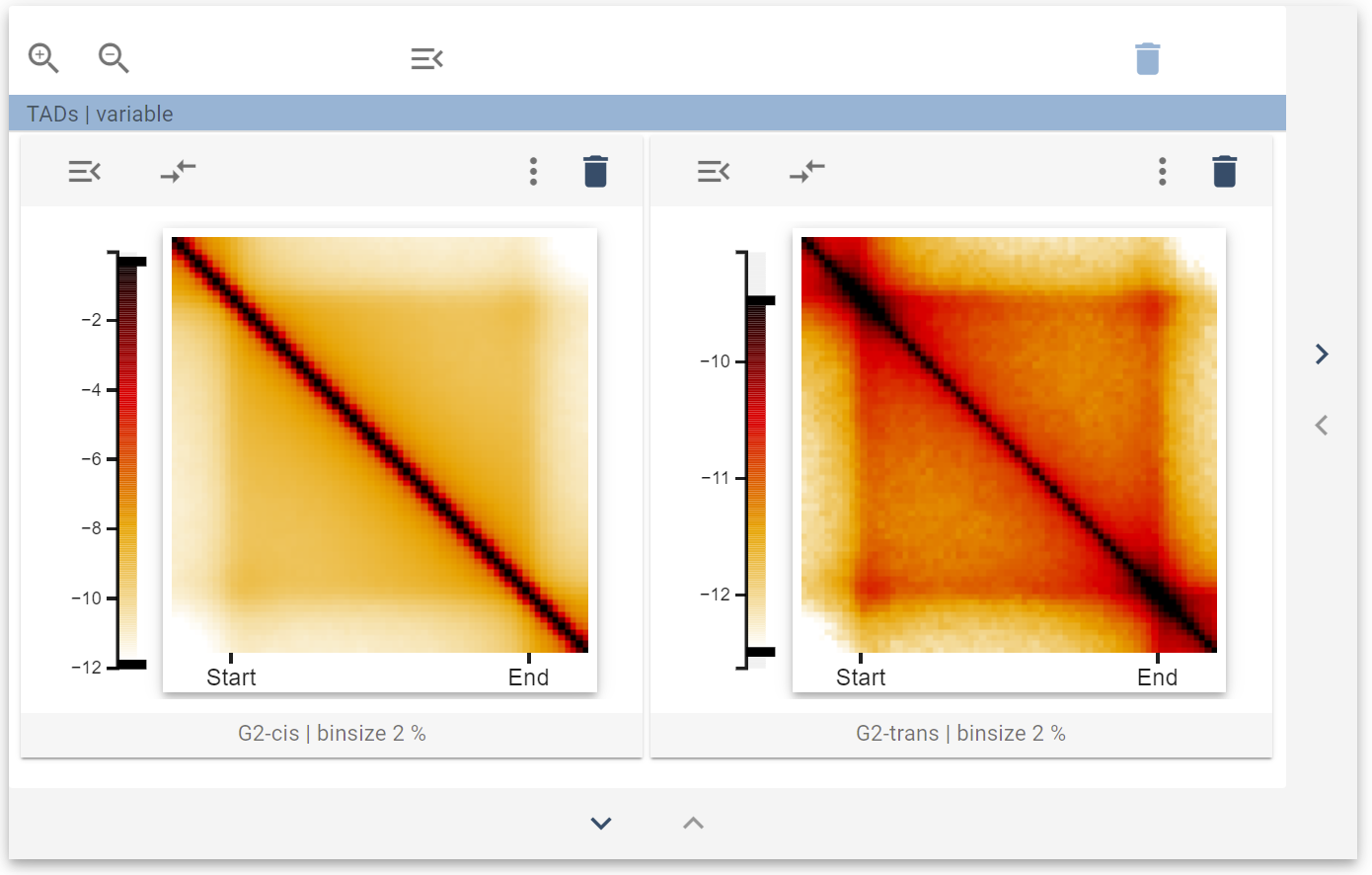
This view now represents the average behavior of cis-sister and trans-sister contacts around TAD-boundaries. If you want to look at the heterogeneity of, e.g., trans-sister contacts around TADs, you can expand the size of your widget collection to add a new slot and fill it with a 2D-embedding widget:
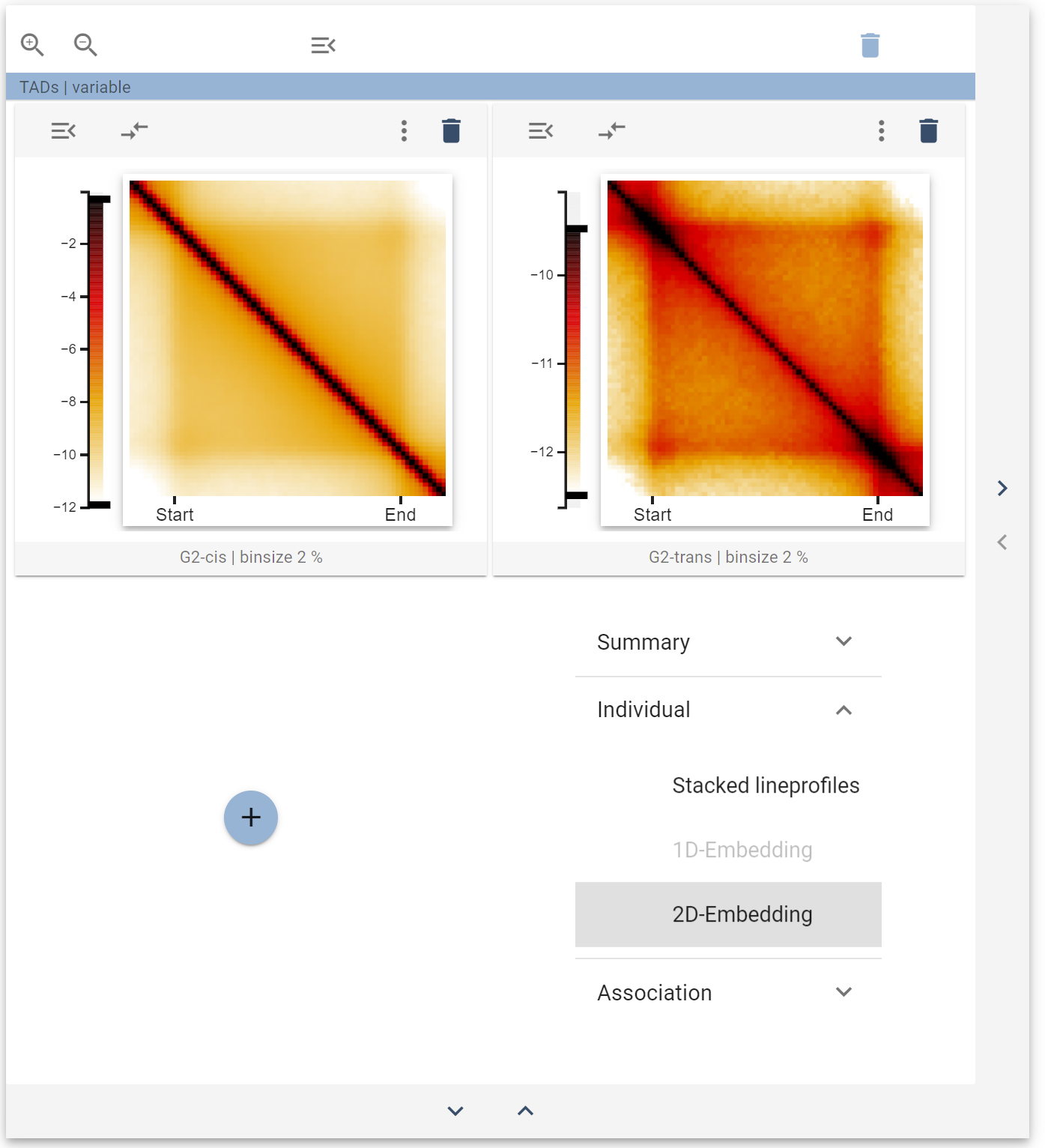
Once you have added the widget, you can select the trans-sister contacts as features to display. Your widget collection should now look like this:

The 2D-embedding widget now allows you to look at the heterogeneity of trans-sister contacts at TADs (see the section about this widget for more details).