Genome assemblies
HiCognition allows managing genome assemblies to be able to work with different assemblies and organisms in parallel.
Adding genome assemblies
To add a genome assembly click the Add Genome Assembly button in the data management drawer:
This will open a dialogue that lets you define a new genome assembly:
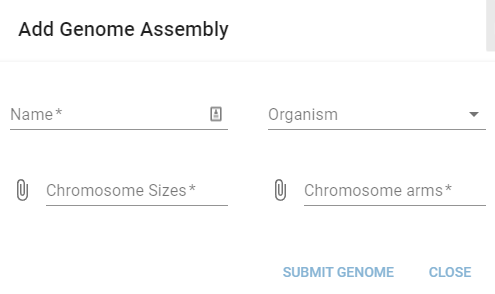
Here you need to give your assembly a name (this needs to be unique), select the corresponding organism, and upload a file specifying the sizes of the chromosomes and the chromosomal arms. The chromosome sizes file needs to define the name, start, and end of each chromosome and use a tab-separator:
Chromosome sizes
chr1 0 1000000
. . .
. . .
. . .
The chromosome arms file needs to define the chromosomal arms also with name, start and end and use a tab-separator:
Chromosome arms
chrom start end
chr1 0 125200000
chr1 125200000 249250621
. . .
. . .
. . .
Here, the two arms are written in separate rows with the same chromosome name as an identifier.
How to create chromosome sizes and chromosome arms?
All you need is pandas and bioframe.
We fetch chromosome sizes and centromeres with bioframe directly from the UCSC database, then we calculate the arms.
chromsizes = bioframe.fetch_chromsizes("hg38")
centromeres = bioframe.fetch_centromeres("hg38")
arms = bioframe.make_chromarms(chromsizes, centromeres)
Two detailed examples of creating these files can be found as a notebook in the HiCogntion repository.
Managing genome assemblies
You can view your genome assemblies by clicking the Show Genomes button in the data management drawer:
This will open a dialogue that lets you view all available genome assemblies:
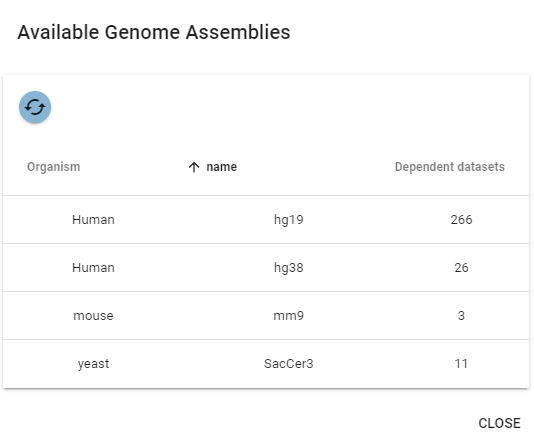
Here, you can look at all the genomes and check how many datasets depend on them.
If you want to delete a genome, first delete all dependent datasets, then you can delete it from the genome table.