Region-set focus approach
Region-set focus approach
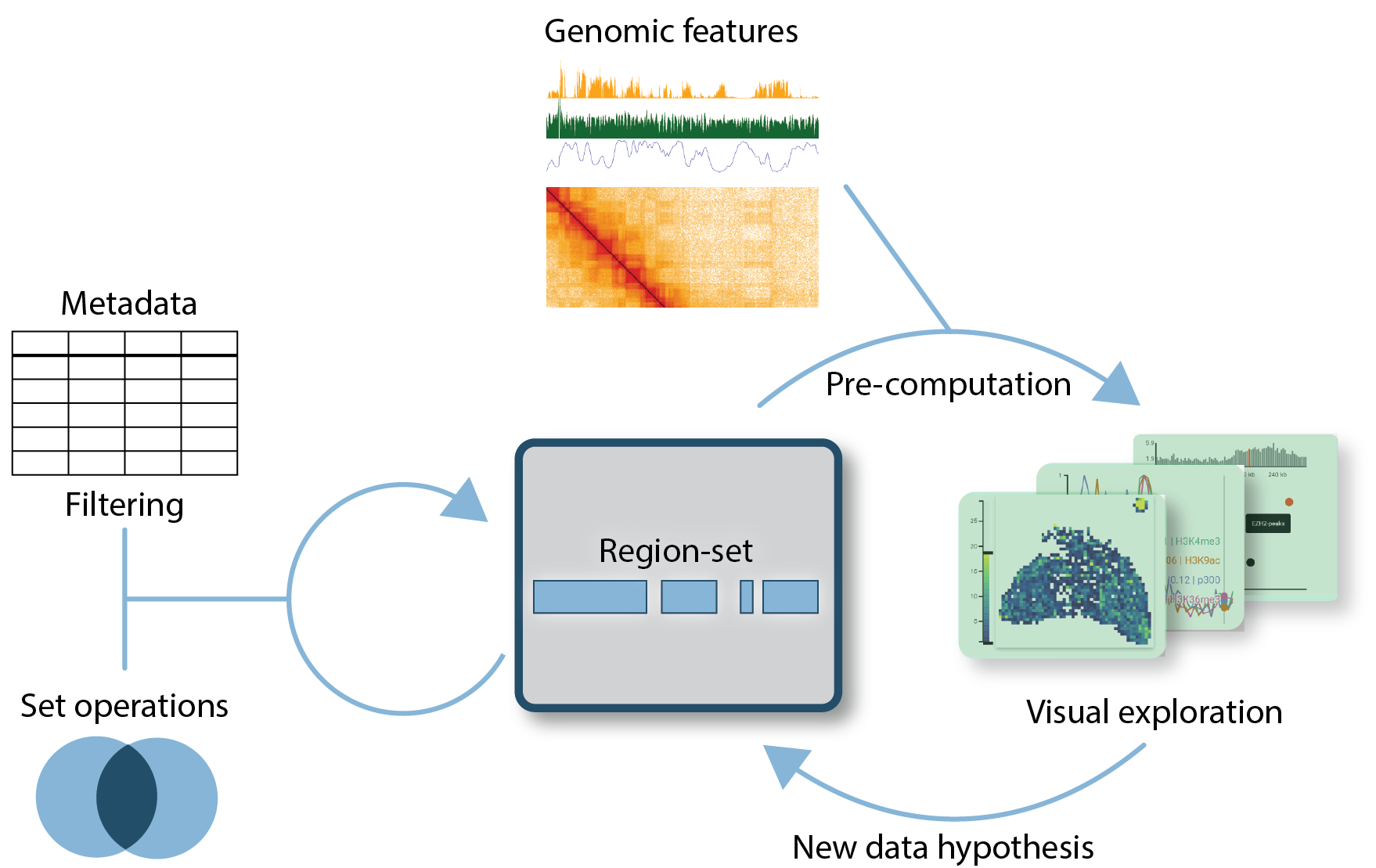
The idea behind the region-set focus approach is that there is a central region-set of particular interest in many biological questions. In contrast, other genomic data is mainly used to find associations. Specifically, a region-set of interest often arises either because the broad biological question gives a natural constraint (e.g., genes/enhancers in the gene expression field), or a particular experimental procedure produces a new region-set that is not characterized, for example, the set of expressed genes or changed 3d-genome interaction patterns in a perturbation condition. Many analysis questions associated with these region-sets of interest can be abstracted into a small number of tasks that are either associated with testing a specific hypothesis or with generating new hypotheses:
- Exploring average behavior: What is the average magnitude of a specific 1d-/2d-genomic signal?
- Exploring heterogeneity: Is the population of regions homogeneous with respect to a collection of 1d- or 2d-features? What subsets have different behavior with respect to a collection of 1d- or 2d-features?
- Enrichment analysis: What other regions are enriched?
With an explicit focus on region-sets of interest, these tasks can be formalized to the degree that allows automated precomputation. Here, the user selects a particular region-set and maps genomic features to them by submitting a precomputation job. The particular pre-computation job is dependent on the type of task and the type of genomic feature, where genomic features can either be continuous 1d- or 2d-signals along the genome (for example, a ChIP-seq signal of a particular protein), discrete signals along the genome (for example the binding sites of a protein of interest) or a collection of the above (e.g., the binding sites of chromatin modifiers). The results of these pre-computations can then be explored using visualization concepts tailored to the feature type.
An implementation of the region-set focus approach would allow users to complement implicit exploration of genomic features in a track-based browser with explicit analyses separated into the tasks mentioned above. Testing a specific hypothesis would allow robust evaluation of average behavior since appropriate pre-computation and visualization tools can be employed. When generating new hypotheses about the data, specialized visualization concepts can help find robust and complex associations that lead to better hypotheses. Additionally, making these analyses explicit allows storing user sessions and sharing them with collaborators, thereby fostering scientific exchange.